Making a Brain Template
antsMultivariateTemplateContstruction.sh.Here’s the entirety of the script so that you have an idea of how it all looks together.
#!/bin/bash
#SBATCH --time=75:00:00
#SBATCH --ntasks=1
#SBATCH --nodes=1
#SBATCH --mem-per-cpu=32768M
#SBATCH -o /fslhome/username/logfiles/dataset/output_temp.txt
#SBATCH -e /fslhome/username/logfiles/dataset/error_temp.txt
#SBATCH -J "ucsdtemp"
#SBATCH --mail-user=LongbranchPennywhistle@gmail.com
#SBATCH --mail-type=BEGIN
#SBATCH --mail-type=END
#SBATCH --mail-type=FAIL
export PBS_NODEFILE=`/fslapps/fslutils/generate_pbs_nodefile`
export PBS_JOBID=$SLURM_JOB_ID
export PBS_O_WORKDIR="$SLURM_SUBMIT_DIR"
export PBS_QUEUE=batch
export OMP_NUM_THREADS=$SLURM_CPUS_ON_NODE
export ANTSPATH=/fslhome/username/bin/antsbin/bin/
PATH=${ANTSPATH}:${PATH}
ROOT=/fslhome/username/
DATASET=${ROOT}/compute/dataset/
TEMPLATE=${DATASET}/template/
cd ${TEMPLATE}
# Construct template
${ANTSPATH}/antsMultivariateTemplateConstruction.sh -d 3 \
-o T_ \
-i 4 \
-c 5 \
-g .25 \
-n 1 \
-r 1 \
-s CC \
-t GR \
-z ${TEMPLATE}T_template0.nii.gz \
${TEMPLATE}*.nii.gz- -i : this is the number of registration iterations that each image contributing to the template undergoes. You should use atleast 4 but be warned that each iteration takes a lot more time than the previous one.
- -c : this controls for parallel computations. 5 indicates a SLURM system. There are also options for SGE qsub, PEXEC, XGrid, and PBS qsub
- -g : this is the gradient step size. The smaller this is the more carefully each iteration builds on the previous one.
- -n : do N4BiasFieldCorrection for all images contributing to the template (0 = off, 1 = 0n). If you’ve already performed this command on the contributing images it’s not necessary but it doesn’t really do any harm to leave this on so I typically do.
- -r : do a rigid-body registration of images before creating demplate (0 = off, 1 = on). This is only useful if you don’t have an initial template that you’re building on
- -s : this describes the similarity metric used to register images
- -t : this is the type of transformation used for registeriing images. In this instance “GR” referes to “Greedy-SyN”
- -z : the target of all the contributing images. If not specified then an unbiased starting point is made by averaging all of the inputs.
: the last space is used for specifying the contributing images. If you look at my script you can see that all of my images are in the same directory. It’s important that all contributing images are in the same directory and this script is executed from there.
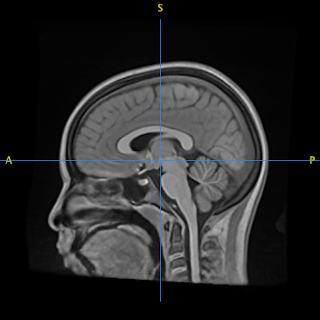
Sagittal view of the resulting template



Hi,
ReplyDeleteI have only 10 MRI scans. I used the same command but got a blurred template. Is there a way to make it sharper?